
Registration Deadline: August 25, 2020
Co-organizers:
Xiaoqian Jiang1 (UTHealth), Genevera Allen2 (Rice University), Devika Subramanian (Rice University),
Assaf Gottlieb (UTHealth), Ioannis Kakadiaris (University of Houston), Yejin Kim (UTHealth)
Sponsor:
1Center for Secure Artificial intelligence For hEalthcare (SAFE)
School of Biomedical Informatics, UTHealth
And
2Center for Transforming Data to Knowledge (the D2K Lab), Rice University
And
Gulf Coast Consortia (GCC) and the GCC cluster of AI in Healthcare
Project Manager:
Marijane Detranaltes (UTHealth)
Steering Committee:
Shayan Shams (UTHealth), Ananth V Annapragada (Texas Children’s Hospital), Kai Zhang (UTHealth)
Architectural Support: Robert III Jolly, David Ha, Luyao Chen, Marcos Hernandez
Logistic Support: Queen Chambliss, Angela Wilkes, Xiaohong Bi
Student Volunteers: Tongtong Huang, Yan Chu





The Kaggle link for this competition, please see the information below
https://www.kaggle.com/c/covid19houstondatathon/overview
The COVID-19 Houston Datathon is an online challenge to predict the regional hospitalization and mortality patterns of COVID-19 in Houston, Texas. This Datathon is jointly organized and sponsored by the Center for Secure Artificial intelligence For hEalthcare at the UTHealth School of Biomedical Informatics, and Data to Knowledge lab at Rice University. Undergraduate, master, and doctoral students from the institutes within the Gulf Coast Consortia (including UTHealth, MDACC, UH, Rice, TAMU, UTMB, IBT, and Baylor) and colleges near TMC are highly encouraged to apply. The event will have up to $1,500 in prizes for the winners. This is an individual-based event (no team participation).
Objective
The goal is to develop a prediction model using local county-level data to estimate the changes in hospitalization and mortality rates in the greater Houston area encompassing 8 counties (Harris, Fort Bend, Montgomery, Brazoria, Galveston, Liberty, Chambers, and Austin) in the state of Texas, USA.
Problem
Accurate and timely prediction of local trends for pandemics will have profound implications to medical resource preparation and policy adjustment evaluation. In this Datathon, we will focus on predicting daily hospitalization cases (COVID-19 general beds + ICU beds) and cumulative mortality cases based on previous observations. We will provide daily hospitalization and mortality statistics (together with infection cases, recovery cases, active cases, test cases) for nine counties in Texas. In addition, we will provide data related to population mobility, demographics, mask usage, which might contain features related to behavioral patterns affecting the transmission.
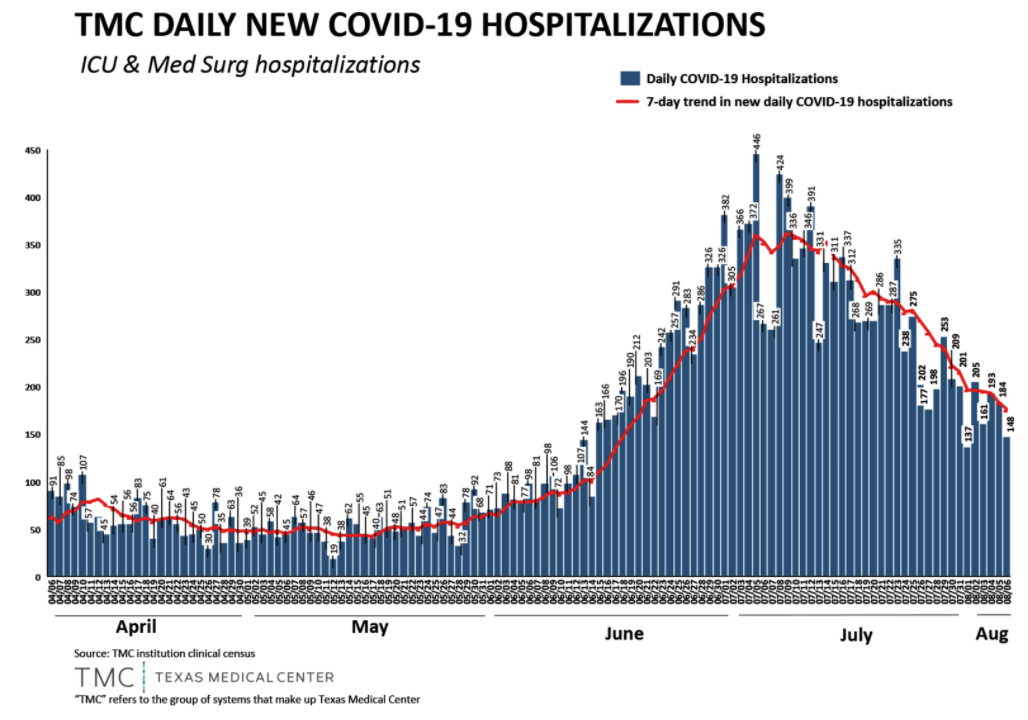
./data/time_series_covid19_confirmed_HOU.csv
Confirmed cases data consists of accumulated confirmed cases at 8 counties in Greater Houston between 04/01/2020 and 09/06/2020. In addition, longitude, latitude, and FIPS are provided, which may serve as foreign keys to query mask survey data.
Confirmed cases data is in a single .csv file (time_series_covid19_confirmed_HOU.csv) with the format below:
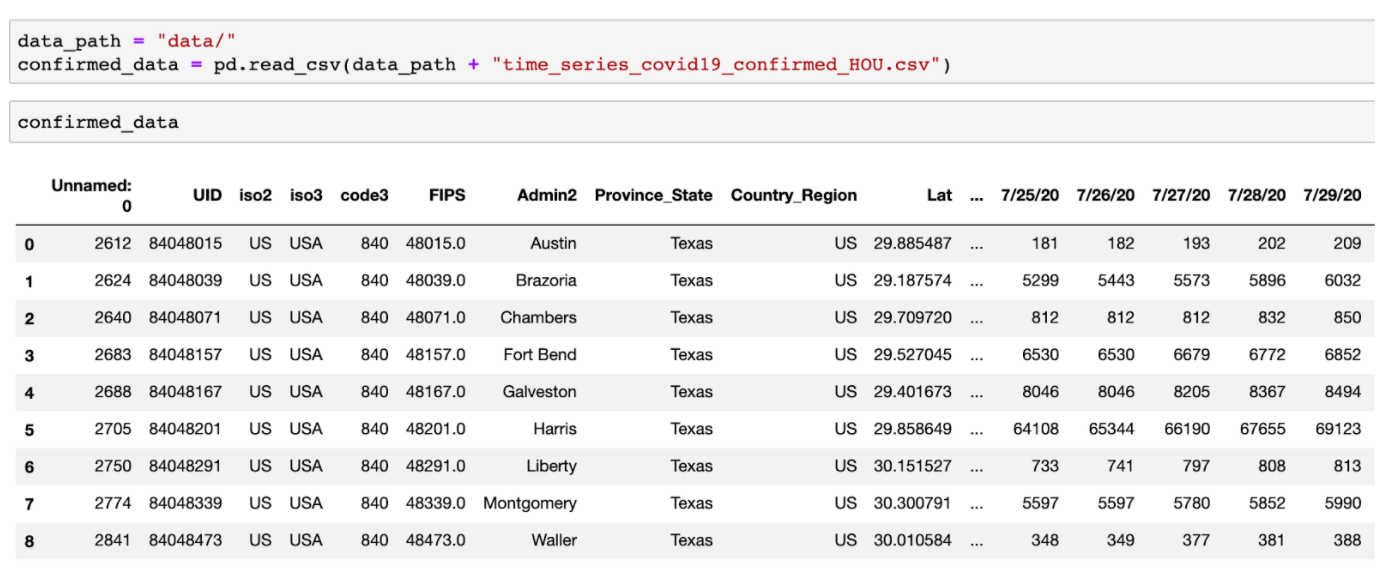
COVID-19 deceased cases data
./data/time_series_covid19_deaths_HOU.csv
Deceased data consists of accumulated deceased cases at 8 counties in Greater Houston between 04/01/2020 and 09/06/2020. In addition, longitude, latitude, and FIPS are provided, which may serve as foreign keys to query mask survey data.
Deceased cases data is in a single csv file (time_series_covid19_death_HOU.csv) with the format below:
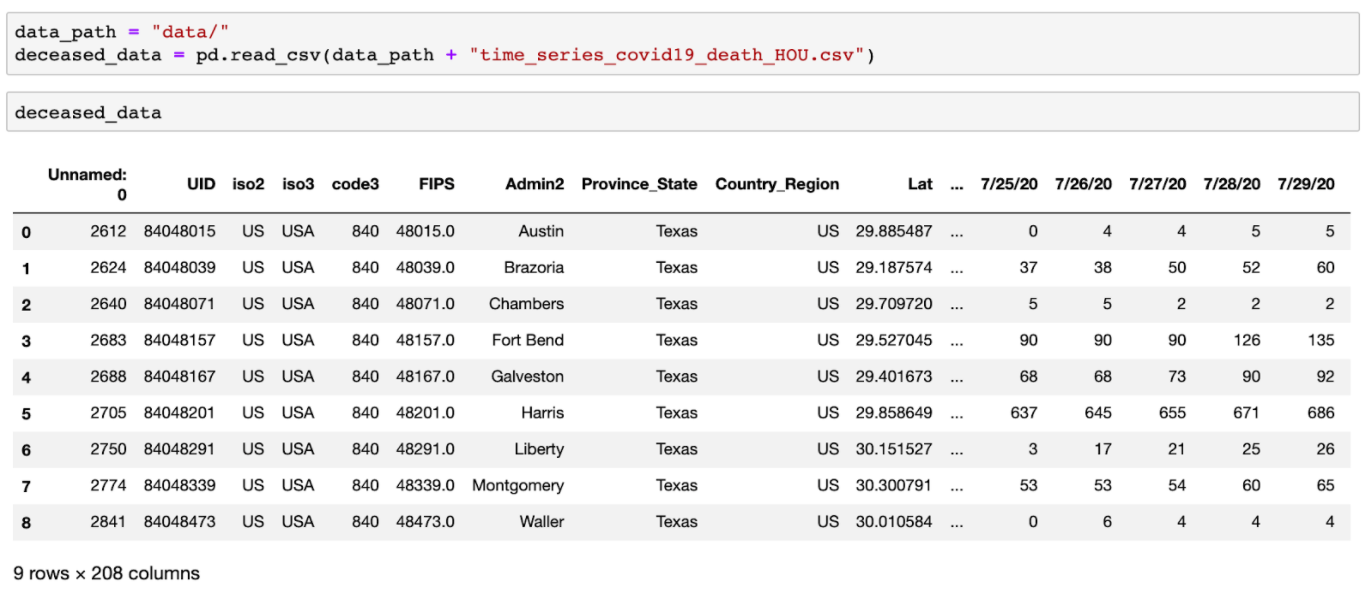
COVID-19 mask usage survey
./data/mask-use-HOU.csv
COVID-19 mask usage survey conducted by The New York Times to estimate the mask usage by county in the United States. Data comes from over 250,000 online interviews between 07/02/2020 and 07/14/2020. Specifically, each interview involves how often the participant wears a mask publicly when he or she expects to be within six feet of another person.
The data includes the following definition:
COUNTYFP: The county FIPS code.
NEVER: The estimated share of people in this county who would say never responding to the question “How often do you wear a mask in public when you expect to be within six feet of another person?”
RARELY: The estimated share of people in this county who would say rarely
SOMETIMES: The estimated share of people in this county who would say sometimes
FREQUENTLY: The estimated share of people in this county who would say frequently
ALWAYS: The estimated share of people in this county who would say always
Mask usage survey data is in a single csv file (mask_use_HOU.csv) with the format below:
COVID-19 Hospitalization data
./data/{county_name}_hosp_{end_date}.xlsx
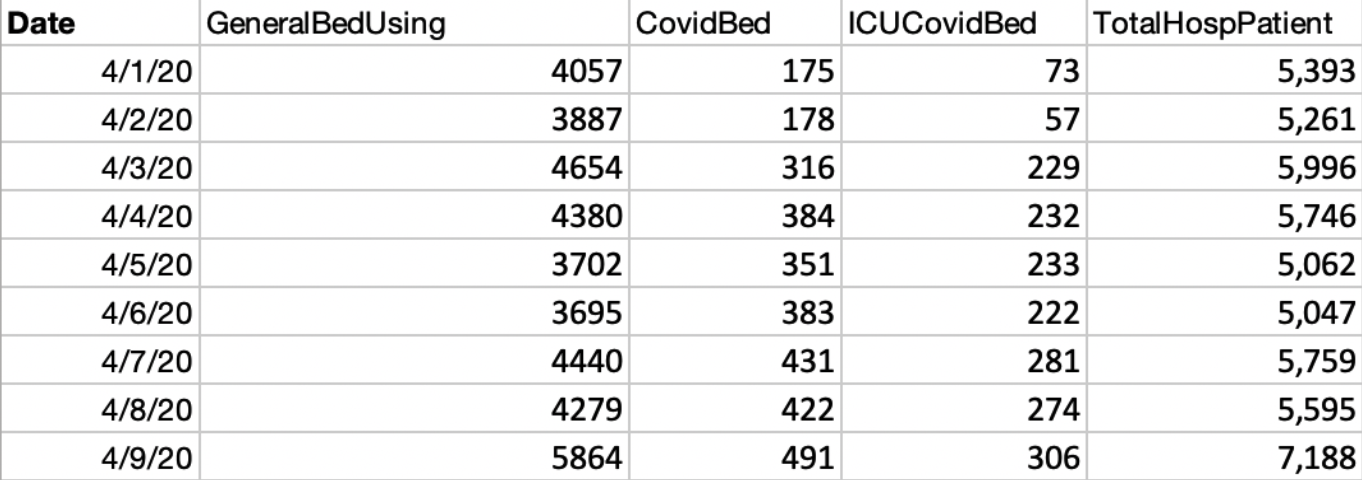
The county-level hospitalization at 8 counties in Greater Houston includes COVID-19 patients in general beds, COVID-19 patients in ICU (no intersection with general bed), total general beds, and total hospitalization patient census. The dataset is available from SETRAC.
Hospitalization data in each county is stored as a separate xlsx file ({county_name}_hosp_{end_date}.xlsx) with the format below:
County FIPS and population data
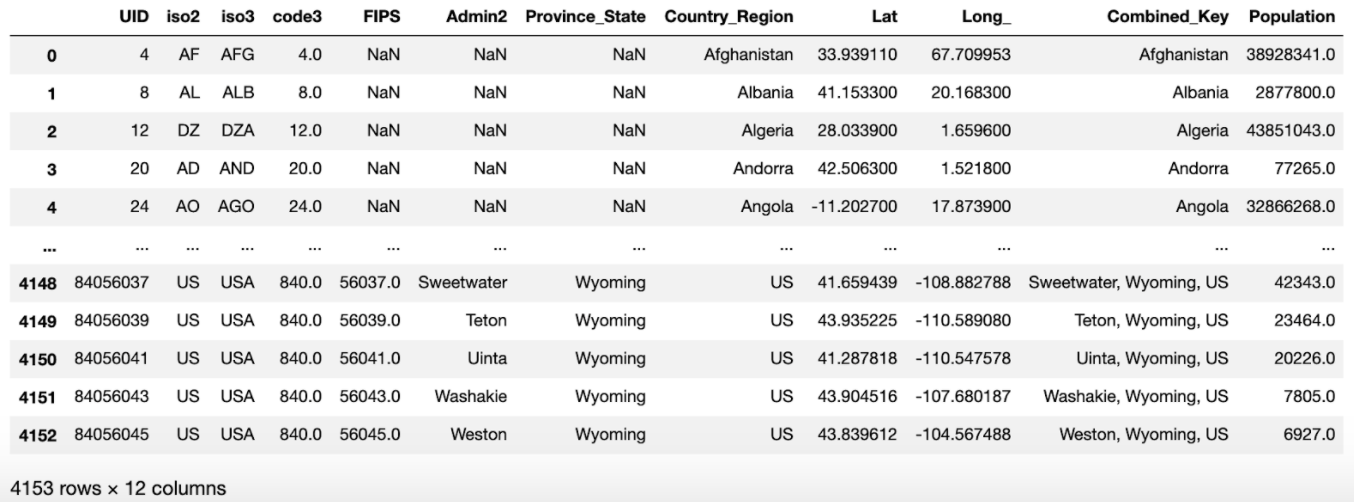
./data/UID_ISO_FIPS_LookUp_Table.csv
FIPS data is used to check county code and population. It’s in a single csv file with the following format:
Leaderboard
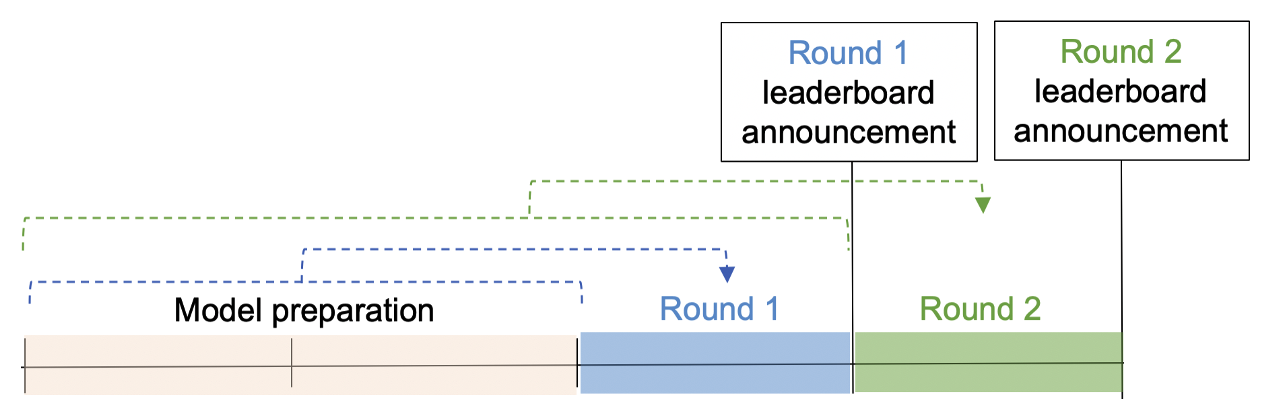
The Datathon will involve two rounds of competition; one for each week after 09/07/2020. The participants will have 2 weeks to prepare and fine-tune their model.
In the first round, the evaluation will use data between 09/07/2020(beginning of the competition) and 09/13/2020 (2 weeks after the start) and top candidates’ performance will be published on a dashboard. Participants should only use data on or before 09/06/2020 to predict the incoming week.
In the second round, participants can update their model and incorporate data from the first period to make predictions for the next week (09/14/2020 - 09/20/2020). Similarly, participants should only use data on or before 09/14/2020. The submitted solutions will be evaluated based on the ranking score (elaborated in the next section).
| Model preparation | 08/26/2020 - 09/06/2020 |
| Round 1 evaluation | 09/07/2020 - 09/13/2020 |
| Round 2 evaluation | 09/14/2020 - 09/20/2020 |
Round 1 Ranking (09/07/2020 – 09/13/2020)
| Rank | ID | Score |
|---|---|---|
| 1 | 0003 | 16 |
| 2 | 0009 | 20 |
| 3 | 0008 | 24 |
| 4 | 0006 | 28 |
| 5 | 0005 | 32 |
| 6 | 0010 | 55 |
| 7 | 0007 | 64 |
| 8 | 0012 | 71 |
| 9 | 0011 | 68 |
| 10 | 0013 | 72 |
| 11 | 0004 | 78 |
| 12 | 0002 | 99 |
| 13 | 0001 | 101 |
Round 2 Ranking (09/14/2020 – 09/21/2020)
| Rank | ID | Score |
|---|---|---|
| 1 | 0008 | 24 |
| 2 | 0006 | 28 |
| 3 | 0010 | 28 |
| 4 | 0009 | 29 |
| 5 | 0005 | 31 |
| 6 | 0003 | 33 |
| 7 | 0001 | 51 |
| 8 | 0007 | 76 |
| 9 | 0004 | 79 |
| 10 | 0014 | 79 |
| 11 | 0011 | 81 |
| 12 | 0012 | 85 |
Combined Ranking (09/07/2020 – 09/21/2020)
| Rank | ID | Score |
|---|---|---|
| 1 | 0003 | 16.5 |
| 2 | 0008 | 20.5 |
| 3 | 0009 | 24 |
| 4 | 0006 | 27 |
| 5 | 0005 | 32 |
| 6 | 0010 | 52 |
| 7 | 0007 | 60 |
| 8 | 0011 | 64 |
| 9 | 0012 | 71 |
| 10 | 0004 | 73 |
| 11 | 0001 | 88 |
Ranking Score Calculation
We will use mean squared logarithmic error (MSLE) of hospitalization and deceased case prediction to evaluate the performance of submitted models on each county. Final scores will be evaluated based on the sum of ranking in each county. We will provide evaluation codes.
MSLE stands for the mean over the observed data of the squared differences between the log-transformed true and predicted values, or writing as a formula:
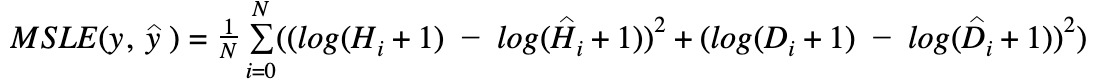
where:
N is the total number of observations
Hi is actual hospitalization value at time i
Ĥi is your hospitalization prediction at time i
Di is actual mortality value at time i
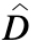 i is your mortality prediction at time i
i is your mortality prediction at time i
In case of equal MSLE scores the leaderboard, we will apply a secondary evaluation metric -- mean squared error (MSE) of hospitalization and deceased case prediction.
MSE stands for the mean over the observed data of the squared differences between the targets and predicted values, or writing as a formula:
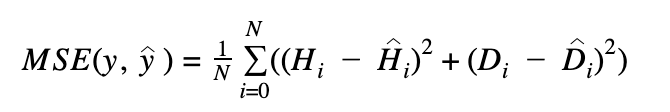
where the meaning of all parameters are the same as above.
Submission
In each competition round, participants are asked to provide predictive hospitalization and mortality cases for the next 7 days. The test/submission file format is identical in both competition rounds. Note that our evaluation metric is independent of Kaggle's default leaderboard ranking settings, so please wait for our final announcement for your correct ranking scores.
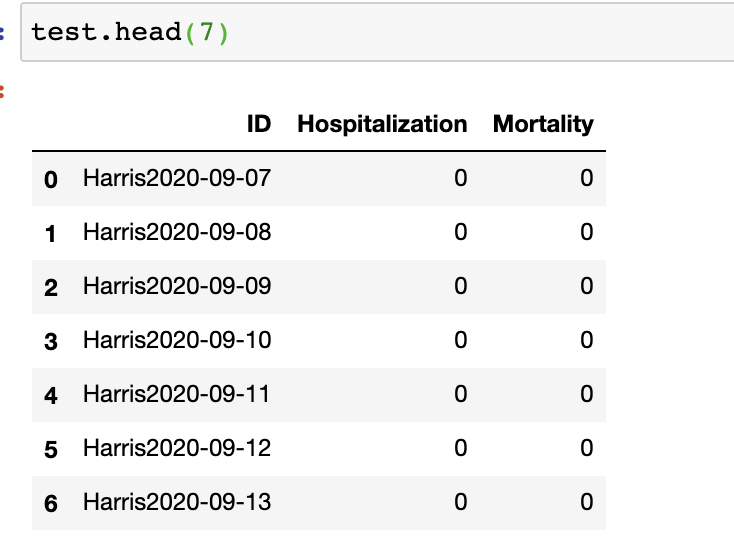
Participants can make predictions with any computational method(s). Test data contains an IDcolumn with format (county_name+date), a hospitalization column, a mortality column that we’d like to compute the error. Note that the date column and county column are necessary as they decide how to match submission results and actual data. The default hospitalization and mortality values in the file are all set as 0. It is a .csv file (test.csv) with the format below:
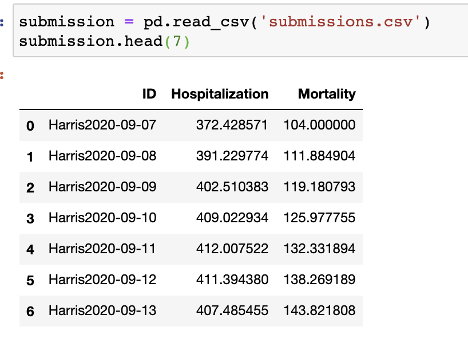
Involving 8 counties (i.e. Harris, Fort Bend, Montgomery, Brazoria, Galveston, Liberty, Chambers, and Austin) in Texas, the submission file should be saved as one csv file (submissions.csv) with format below:
A total of $1,500
Institution Specific Prizes:
In addition, participating students will receive suvanariors sponsored by the GCC and the GCC cluster of AI in Healthcare
Find additional frequently asked questions (FAQs) and answers here:
https://docs.google.com/document/d/1k1yJu7igk2uwUWde4FmKwN1dN-vrqGglBHzAgWIEWUo
