
The 2nd SBMI Healthcare Machine Learning Datathon is calling capable and motivated undergraduate and graduate students from Gulf Coast Consortia institutions and other Houston area universities. Come join us for this great opportunity to challenge your coding skills, meet new people, and enjoy the gathering of young hackers. This 24-hour Datathon is organized by the Center for Center for Secure Artificial intelligence For hEalthcare(SAFE) at the School of Biomedical Informatics in UTHealth. The event is sponsored by Vir Biotechnology, for a total prize of $2,100 for the winners. Undergraduate, master, and doctoral (only 1st and 2nd year) students from the institutes within the Gulf Coast Consortia (include UTHealth, MDACC, UH, Rice, TAMU, UTMB, IBT, and Baylor) and colleges in the vicinity of TMC are highly encouraged to apply. This is individual-based event (no team participation).
Identifying new treatment for diseases is the long-standing goal of medicine. The cost of drug discovery is very high in the traditional setting. Systematic integration of previous results and knowledge might change the game by identifying highly promising drugs to save cost and speedup discovery.
High-throughput drug screening using computational approaches has the potential to substantially improve cost-efficiency by automatically estimating drug sensitivity based on genomic and pharmacological data.1–7 These computational methods utilize drug sensitivity data at certain cell lines and predict promising drugs that potentially have high sensitivity in other cell lines. In this Datathon, participants are asked to build a prediction model that ranks promising drugs in given cancer cell lines.
Key words: drug repositioning, collaborative filtering, recommender system, cold start, graph convolution neural network, tensor factorization, random walk.
Objective
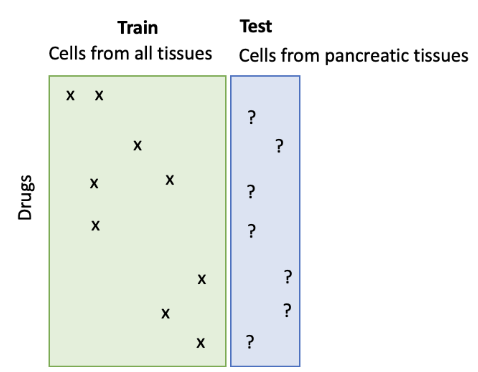
The goal is to use machine learning to predict drug’s sensitivity in given cell lines. Participants should rank drugs that are likely to be sensitive (i.e., relative inhibition > 50%) in cell lines given in test sets. A key challenge is that testing cell lines have limited experimental drug response data in the training set. Participants are encouraged to use relative dense observations from other types of cancer cell lines to predict these data-rare cancer cell lines.
Cold-start problem
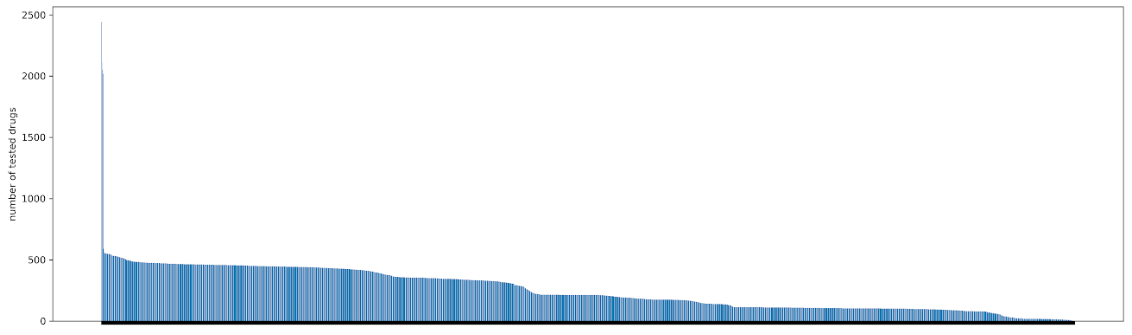
A key challenge lies in the imbalances of experimental data for different tissues. For example, we have accumulated many drug sensitivity experimental data for major tissues’ cell lines (such as lung and breast). Traditional methods target at studying commonly observed tissues, 8–15 which take known drug response data at certain cell lines and attempt to find other drugs responses at other cell lines within the data-rich tissues.
In this Datathon, we will focus on ranking drugs for pancreatic cancer, which is deadly and incurable. The study on pancreatic cancer cell lines is limited and training data for machine learning models is insufficient. A potential way to mitigate this data paucity problem is to utilize common features - gene expression level, because these different tissues share biological commonality partly in terms of gene expression and therefore respond drugs in similar ways.16 We will provide the cell’s gene expression level as external features, and participants need to incorporate the features into the prediction model.
| ./train.csv |
Train data is a drug’s sensitivity of given pair of (drug,cell). We will use relative inhibition (RI) as a drug sensitivity measure. The RI has been binarized as 1 if the drug shows efficacy in the cell; and 0 otherwise.
The train data are in a single csv file (train.csv) or binarized pickle file (train.pkl) with format below:
| ./drug/target_genes.tsv ./drug/fingerprint.tsv (optional) ./drug/profiles.csv (optional) |
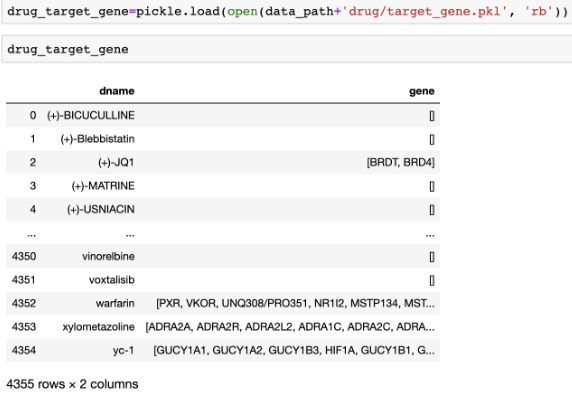
One of the most important features for drug repositioning is drug’s target genes. Drugs are suppressing specific target genes, which consequently causes modification of the diseases. We will provide drug’s features as tsv files and binarized pickle files).
(Optional) Additionally, one can further investigate the drug’s chemical features using molecular structure. Contestants can freely use Molecular ACCess System (MACCS) fingerprints17 and native chemical compounds to boost prediction performance. MACCS fingerprints contains 166 chemical structures such as the number of oxygens, S-S bonds, ring. In addition, we represented drug as a native chemical structure using SMILES. SMILES is a linear notation to represent chemical compound in a unique way; in the SMILES representation atoms are represented as their atomic symbols (e.g., c for carbon); special characters are also used to represent relationship (e.g., “=”: double bonds; “#”:triple bonds; “.”:ionic bond; “:”: aromatic bond)18. SMILES can provide richer features space that strictly represent functional substructures and express structural differences such as compound’s chirality19.
| ./cell/gene_expressions.csv ./cell/profiles.csv (optional) |
The cell lines come from 14 different tissues including lung, ovary, and skin and 14 different types of cancer including carcinoma, adenocarcinoma, and melanoma. We provided gene expression profiles of each cell line using Fragments Per Kilobase of transcript per Million reads mapped (FPKM).20,21 This gene expression profiles is an accurate quantification of the cell’s genetic status. Note that some cell lines have missing gene expression.
(Optional) We also provide cell line’s profiles with identifiers linking external databases.
Contestants are asked to select the most efficacious 20 drugs for each 44 pancreatic cancer cell lines given in the test set. Contestants should predict and rank the efficacious drugs with computational method. Test data contains drug and cell line that we’d like to predict the sensitivity. It is a csv file (test.csv) with format below:
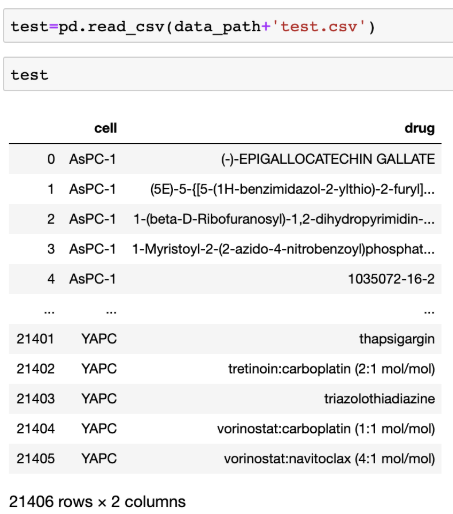
The submission file (submission.csv) should be formatted as:
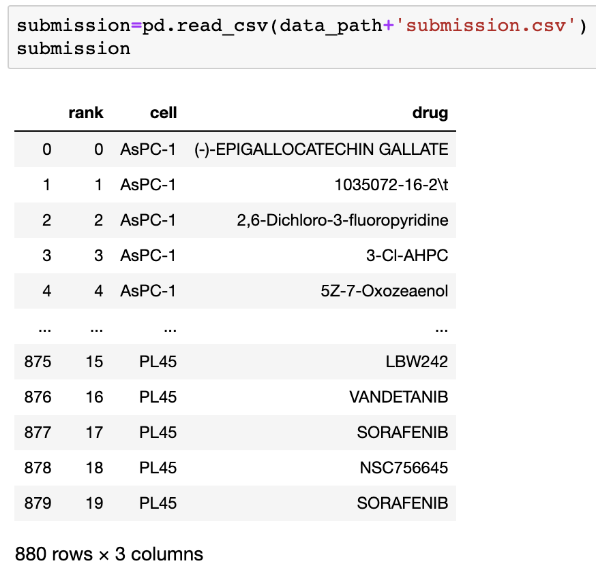
The lowest rank (0, 1, 2, 3, ...) correspond to the most efficacious drugs. Submissions will be judged on the ranking scores - normalized discounted cumulative gain (NDCG).
As our ultimate goal is to rank drugs based on likelihood of efficacy and help prioritizing drug experiments in vitro, we will measure the accuracy of the model as ranking performance. For ranking measure, we will evaluate normalized discounted cumulative gain (NDCG). In information retrieval, cumulative gain is the sum of relevance values (i.e., relative inhibition) of high-ranked drugs per each query (cell). Discounted cumulative gain (DCG) is the sum of graded/weighted/discounted relevance scores in the top ranking list. The formula for DCG accumulated at top-20 ranking list is
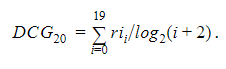
DCG will be high if highly efficacious drugs are ranked high and/or if highly efficacious drugs are ranked higher than marginally efficacious drugs. The DCG score can be normalized by the maximum DCG or ideal DCG (IDCG) in which the ranking is perfectly matched:
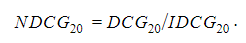
We will use averaged NDCG20 for all 44 cell lines as a final performance measure of this Datathon.
Useful links and literature:
First prize: $1200
Second prize: $600
Third prize: $300
Location:
School of Biomedical Informatics (SBMI), University of Texas Health Science Center at Houston (UTHealth)February 1, 2020
February 2, 2020
Reference:
