
The 1st SBMI Healthcare Machine Learning Hackathon has successfully finished. The first place winner was Rice University PhD student, Qiang Zhang. Bishal Lamichhane, another Rice University PhD student and Carroll Vance, a University of Houston Computer Science undergraduate student, earned second and third place respectively. Thanks for all the dedication and participation. We hope to see you again at the next Hackathon. For more pictures, click here.
The 1st SBMI Healthcare Machine Learning Hackathon is calling for the participation of capable, motivated undergraduate and graduate students from Gulf Coast Consortia institutions and other Houston area universities. Come join us for this great opportunity to challenge your coding skills, meet new people, and enjoy interacting with other young hackers! This 24-hour Hackathon has been organized by the Center for Health Security and Phenotyping (CHSP) at the UTHealth School of Biomedical Informatics and the Texas Institute for Restorative Neurotechnology. The very first event of our Hackathon series is sponsored by Elimu Informatics, Inc., which is providing a $1,200 prize to the winner, along with some great swag for participants.
Annually, approximately 3,000 people in the United States die from Sudden Unexpected Death in Epilepsy (SUDEP) because of a shutdown in brain, cardiac, and breathing function. One way to potentially determine risk is the accurate identification of certain electroencephalographic (EEG) features after epileptic seizures. Unfortunately, human visual analysis of such features is unreliable.
The theme of this Hackathon is to compete on the development of algorithms to detect the end of postictal generalized EEG suppression (PGES) by identifying the onset of slow activity after a generalized tonic-clonic (GTC) seizure. Participants should detect the commencement of slow activity no later than a few seconds after onset. The input consists of signals from multiple channels monitoring the electrical activity in various regions of the brain. Note that there is a lot of noise due to the environment (e.g., patient motion, bed relocation, etc.); also, muscles complicate input via artifacts that are irrelevant to the brain. Experts have expended a good deal of effort to identify and monitor a number of critical patterns, which often requires considering multiple channels.
In this machine learning challenge, we ask the participants to build models (in a justifiable manner) and evaluate final performance, based on accuracy; additional consideration will be given to model interpretability, should participant performance be tied. See this for more details on the detection of PGES.
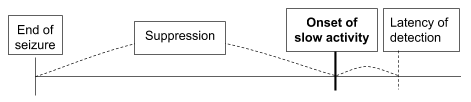
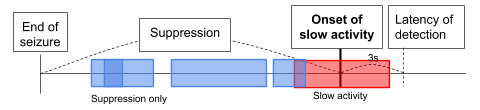
Objective. The objective of this Hackathon is to detect the onset of slow activity after a seizure (Fig. 2). The detection of onset should be done no later than 3 seconds (i.e., latency) after actual onset. Note that detecting onset after the latency period would be trivial in a real-time detection scenario.
Signal. For the purposes of this competition, we will focus on EEG signals, which is key to observing brain activity. The EEGs are sampled from multiple electrodes at a sampling rate of 200 Hz, which captures temporal and spatial patterns of the brain (Fig 3). In this competition, we will only focus on 13 electrodes (Fp1, Fp2, O1, O2, F7, F8, T7, T8, P7, P8, Fz, Cz, Pz), as well as pairwise offsets of two adjacent electrodes (i.e., a so-called montage).
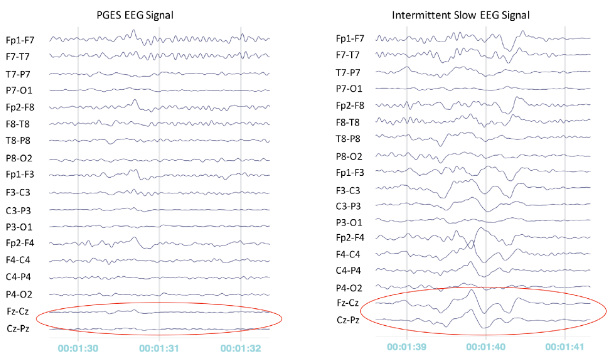
Typical patterns of suppression and slow activity are shown in Fig. 4. Generally speaking, suppression patterns are stable, with no apparent fluctuation (Fig. 4 left); whereas, a slow activity pattern shows moderate fluctuation (Fig. 4 right).
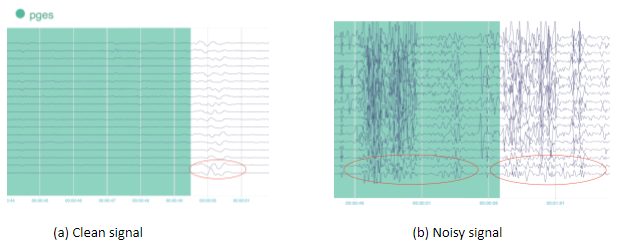
Physicians usually review the EEG signals retrospectively, manually annotating the onset of slow activity. However, EEG signals can be very noisy and there are multiple sources of contamination (Fig. 5), such as heart activity, eye movements, and facial muscle movement, etc. Go here for more details. In consequence, we need an automated tool to support physicians in effectively handling noisy signals in order to derive useful patterns that will enable detection of the onset of slow activity.
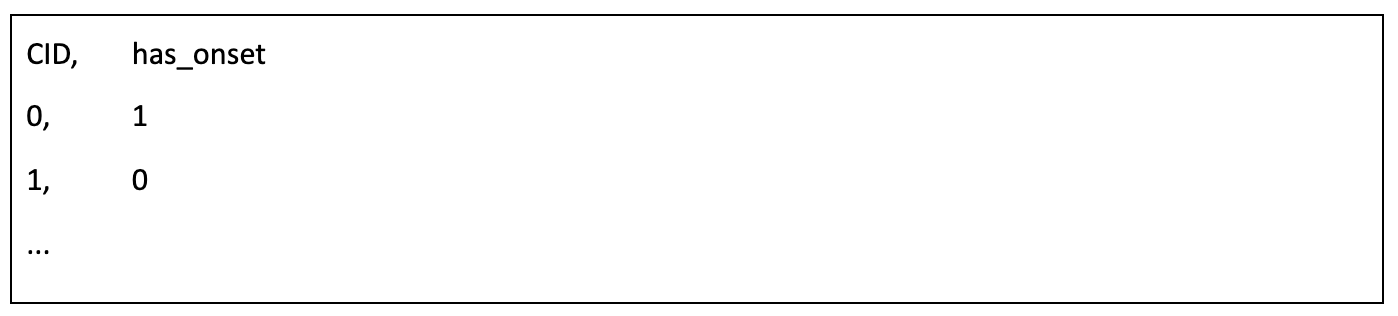
Submissions are judged on the accuracy of detection. Participants must identify whether given short segments of EEG signals (i.e., clips) indicate the onset of slow activity (Fig. 6). Clips in the test file are variable in length. The slow activity clip does not include slow activity signals beyond 3 seconds after onset. Participants should submit a solution file in a CSV format with classification for each test clip. The file should contain a header and have the following format:
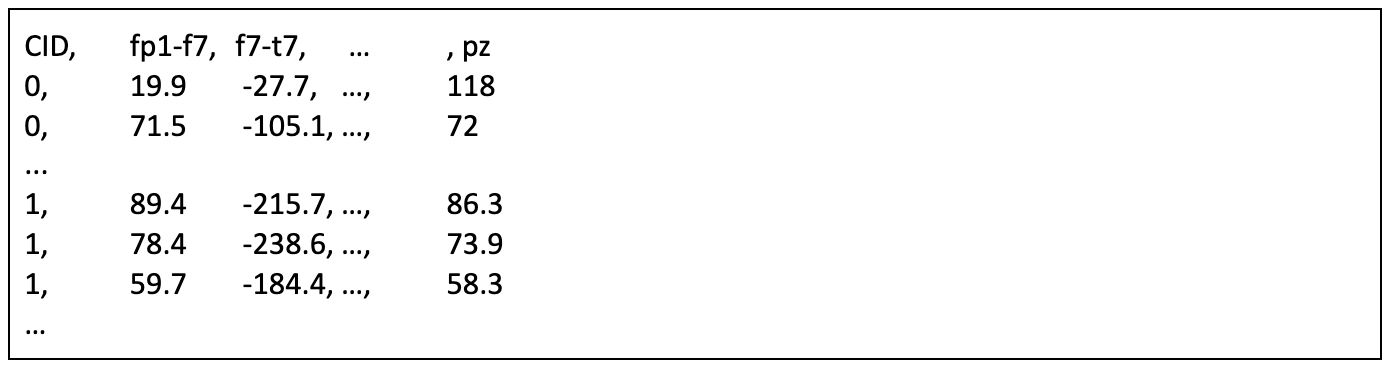
Train data are in a single csv file (train.csv) with format below:
For each patient, the signals have the aforementioned 13 electrodes (fp1, fp2, …, pz) and 10 precalculated pairwise offsets (fp1-f7, f7-t7, …, cz-pz) for reference. Label==1 refers to the after slow activity and label==0 refers to before slow activity. The signals were truncated to the first 180 seconds after the seizure ends; the onset of slow activity primarily occurs during this period.
Test data are in a single csv file (test.csv) with the format below:
Location:
School of Biomedical Informatics (SBMI), University of Texas Health Science Center at Houston (UTHealth)Please note: Contestant on the leaderboard will be invited to submit paper with us to a special issue of BMC Medical Informatics and Decision Making
September 14, 2019
September 15, 2019
Winner: $1,200
RELATED LITERATURE:
