Wednesday, October 8th, 2014
Tens of thousands of dollars: that’s what it cost less than a decade ago to sequence genomes. Today, the cost is around $5,000 and expected to continue to drop.
With genomic analysis of individual patients now so much more affordable, and therapies targeted to specific molecular aberrations being tested in clinical trials, personalized medicine is here. There are currently over 800 ongoing targeted therapy trials at MD Anderson Cancer Center (MDACC) alone.
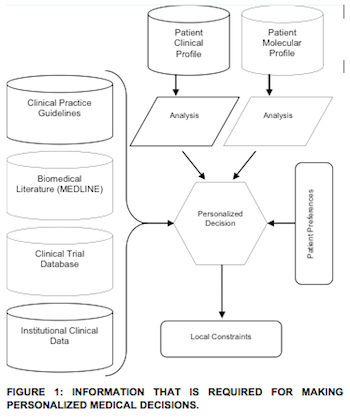 All practitioners hoping to personalize therapy for their patients are facing the same problem: there is simply too much information about the specific patient and the available therapies, all of which must currently be managed manually (see Figure 1).
All practitioners hoping to personalize therapy for their patients are facing the same problem: there is simply too much information about the specific patient and the available therapies, all of which must currently be managed manually (see Figure 1).
While we finally have the capability to deliver personalized cancer therapy, even specialized clinicians need help to interpret genomic data. The rapidly expanding body of biomedical literature is simply too much for anyone to keep up with.
At MDACC, the Institute for Personalized Cancer Therapy (IPCT) facilitates molecular testing and personalized therapy selection.
To recommend a treatment based on a patient’s molecular profile requires many steps: genomic data annotation, literature review to identify molecular aberrations that “drive” the disease process, as well as associations between genomic profiles and specific targeted therapies. This information is used to optimally match patients to approved drugs and open clinical trials of investigational molecular therapies.
The current, manual process of sifting through such a massive corpus of data is inefficient, cumbersome and does not scale. Thus, it cannot support routine (“standard of care”) personalized therapy.
Automated (or at least semi-automated) informatics tools are needed to personalize therapy for all cancer patients. In other words, personalized medicine is a challenge that informatics can address.
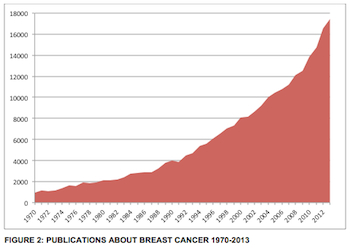 This is a particular problem in breast cancer. Clinically significant, actionable information is continually being discovered (see Figure 2) and is being published at an ever-increasing rate. To identify the knowledge that is clinically actionable, and then to apply it to the care of individual patients, even the most diligent clinician needs help.
This is a particular problem in breast cancer. Clinically significant, actionable information is continually being discovered (see Figure 2) and is being published at an ever-increasing rate. To identify the knowledge that is clinically actionable, and then to apply it to the care of individual patients, even the most diligent clinician needs help.
At SBMI and MDACC, we are developing tools that automatically mine the biomedical literature to identify drugs that can be given to patients who have tumors with specific molecular abnormalities. This information is reviewed by personalized cancer therapy experts and published online (http://personalizedcancertherapy.org) so that this information is available to everyone affected by cancer.
This work is supported in part by the National Center for Advancing Translational Sciences (grant UL1 TR000371), Center for Clinical and Translational Sciences, National Cancer Institute 1U01 CA180964, the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy, the Nellie B. Connally Breast Cancer Research Chair Funds, the MD Anderson Cancer Center (support grant P30 CA016672), and a Cancer Prevention Fellowship (National Cancer Institute grant R25T CA57730).
This is just one more way SBMI is making a difference in medicine through collaborative partnerships and the application of informatics to health care.
written by Elmer Bernstam, MD, MSE
contributions from W. Jim Zheng, PhD and Funda Meric-Bernstam, MD

Dr. Elmer Bernstam, MD, MSE is the Associate Dean for Research and a Professor at UTHealth School of Biomedical Informatics. He is a fellow of the American College of Physicians and the American College of Medical Informatics and is also board-certified in internal medicine and continues to practice. His research interests are in information retrieval, consumer informatics, and clinical decision support.