| MSEA: Mutation Set Enrichment Analysis |
MSEA identifies cancer driver genes by detecting groups of somatic mutations, aka hotspots. |
- Faculty/PI Name: Peilin Jia
- Publications: Jia P, Wang Q, Chen Q, Hutchinson K, Pao W, Zhao Z (2014) MSEA: detection and quantification of mutation hotspots through mutation set enrichment analysis. Genome Biology 15(10):489
- Website Link: https://github.com/bsml320/MSEA
|
|
| VarWalker |
VarWalker performs mutation network analysis of putative cancer genes from next-generation sequencing data. |
- Faculty/PI Name: Peilin Jia
- Publications: Jia P, Zhao Z (2014) VarWalker: Personalized Mutation Network Analysis of Putative Cancer Genes from Next-generation Sequencing Data. PLoS Computational Biology 10(2): e1003460
- Website Link: https://bioinfo.uth.edu/VarWalker.html
|
|
| GATHER |
It is a Gene Annotation Tool to Help Explain Relationships. |
|
|
| SIGNATURE |
It is a web-based resource that simplifies gene expression signature analysis by providing software, data, and protocols to perform the analysis successfully. |
- Faculty/PI Name: Jeffrey Chang
- Publications:
Chang JT, Gatza ML, Lucas JE, Barry W, Vaughn P, and Nevins JR. "SIGNATURE: A Workbench for Gene Expression Signature Analysis." BMC Bioinformatics 12(443), 2011
- Website Link: https://uth.tmc.edu
|
|
| TREC Precision Medicine |
An information retrieval (IR) tool for finding relevant precision medicine scientific literature and clinical trials for specific cancer patients. |
- Faculty/PI Name: Kirk Roberts
- Publications:
- Roberts K, Demner-Fushman D, Voorhees E, Hersh WR, Bedrick S, Lazar A, Pant S. (2017). Overview of the TREC 2017 Precision Medicine Track. Proceedings of the Text Retrieval Conference.
- Roberts K. Automatic Identification of Cancer Precision Medicine Literature Articles. In Submission.
|
|
| Cancer FrameNet |
A natural language processing (NLP) information extraction tool for frame-based information about cancer written in clinical text. |
- Faculty/PI Name: Kirk Roberts
- Publications:
- Roberts K, Si Y, Gandhi A, Bernstam EV. (2018). A FrameNet for Cancer Information in Clinical Narratives: Schema and Annotation. Proceedings of the Language Resources and Evaluation Conference.
- Si Y, Roberts K. A Frame-Based NLP System for Cancer-Related Information Extraction. In Submission.
|
|
| Epiphanet |
EpiphaNet is an interactive knowledge discovery system, which enables researchers to explore visually sets of relations extracted from MEDLINE using a combination of language processing techniques. |
- Faculty/PI Name: Trevor Cohen
- Publications:
- Cohen, T., Whitfield, G. K., Schvaneveldt, R. W., Mukund, K., & Rindflesch, T. (2010). EpiphaNet: An Interactive Tool to Support Biomedical Discoveries. Journal of Biomedical Discovery and Collaboration, 5, 21–49.
|
|
| CLAMP: Clinical Language Annotation, Modeling, and Processing Toolkit |
CLAMP is a comprehensive clinical Natural Language Processing (NLP) software that enables recognition and automatic encoding of clinical information in narrative patient reports. |
- Faculty/PI Name: Hua Xu
- Publications:
- Ergin Soysal, Jingqi Wang, Min Jiang, Yonghui Wu, Serguei Pakhomov, Hongfang Liu, Hua Xu. CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines. JAMIA, Doi: 10.1093/jamia/
- Jun Xu*, Hee-Jin Lee*, Zongcheng Ji*, Jingqi Wang, Qiang Wei, and Hua Xu. UTH_CCB System for Adverse Drug Reaction Extraction from Drug Labels at TAC-ADR 2017. Proceedings of TAC, 2017. (* denotes equal contribution)
- Website Link: https://clamp.uth.edu/
|
|
| SBMI Data Service |
SBMI Data Service provides technical assistance and consulting services to qualified clients about the health data sets available at the school. |
- Faculty/PI Name: Hua Xu
- Website Link: SBMI Data Service
|
|
| R, SAS |
We developed computer codes using R and SAS for various projects in cancer research and other fields. |
- Faculty/PI Name: Dejian Lai
- Publications:
A. Recent Projects in Cancer Research:
- Early Life Exposures to Air Toxics and Risk of Early Childhood Leukemia (2018)
- SPATIAL ANALYSIS OF AMBIENT BENZENE AND CANCER INCIDENCE RATES IN TEXAS (student thesis, graduated in 2017).
- Longitudinal Study of Melatonin, Cortisol and risk of Colorectal Cancer (proposal submitted in 2017)
- Hazardous Air Pollutants and Lymphohematopoietic Cancer Incidence in Houston (NIH funded project).
- USING SPATIAL LINEAR MODELS WITH SAR AND CAR STRUCTURE TO EXAMINE TEXAS LUNG CANCER INCIDENCE RATES (student thesis, graduated in 2017)
B. Some Cancer Related Publications in Last Five Years:
- Symanski E, Lewis PGT, Chen TY, Chan W, Lai D, Ma XM: Air Toxics and Early Childhood Acute Lymphocytic Leukemia in Texas, A Population Based Case Control Study. 2016, Environmental Health: A Global Access Science Source. 2016, Vol. 15, No. 70
- Tong L, Ahn C, Symanski E, Lai D, Du XL. Relative Impact of Earlier Diagnosis and Improved Treatment on Survival for Colorectal Cancer: A US database Study among Elderly Patients. Cancer Epidemiology. 2014, Vol. 38, 733-740.
- Tong LY, Ahn C, Symanski E, Lai D, Du XL: Temporal Trends in the Leading Causes of Death among a Large National Cohort of Patients with Colorectal Cancer from 1975 to 2009 in the United States. Annals of Epidemiology. 2014, Vol. 24, 411-417
- Tong LY, Ahn C, Symanski E, Lai D, Du XL: Effects of Newly Developed Chemotherapy Regimens, Comorbidities, Chemotherapy-related Toxicity on the Changing Patterns of the Leading Causes of Death in Elderly Patients with Colorectal Cancer. Annals of Oncology. 2014, Vol. 25, 1234-1242.
- Wang GD, Lai D, Burau K, Du XL: Potential Gains in Life Expectancy from Reducing Heart Disease, Cancer, Alzheimer's Disease, Kidney Disease or HIV/AIDS as Major Causes of Death in the USA. Public Health. 2013, Vol. 127, 348-356.
|
|
| Machine Learning tools for Longitudinal Brain Connectivity |
Methods to identify neuroplasticity patterns in brains are of the utmost importance in understanding and potentially treating diseases. Diffusion tensor imaging (DTI) allows for an in-vivo estimation of the structural connectome inside the brain and may serve to quantify degenerative process before the appearance of clinical symptoms. We have developed novel machine learning-based strategies to compute longitudinal structural connectomes that allow the discovery and quantification longitudinal patterns. |
- Faculty/PI Name: Luca Giancardo
- Publications:
- Giancardo, L.*, Ellmore, T. M., Suescun, J., Ocasio, L., Kamali, A., Riascos-Castaneda, R. & Schiess, M. C. Longitudinal Connectome-based Predictive Modeling for REM Sleep Behavior Disorder from Structural Brain Connectivity. Proceeding SPIE Med. Imaging (2018).
|
|
| Machine learning-based image (and video) segmentation |
We develop and adapt pipelines for image and video segmentation. These pipelines can be adapted to the multiple use cases by leveraging machine learning techniques that learn by examples. These tools allow for high throughput analysis quantitative analysis of large dataset. We have experience with optical images, MRI and videos. |
- Faculty/PI Name: Luca Giancardo
- Publications:
Here are some examples of new image segmentation pipelines developed
- A Fazlollahi*, F Meriaudeau, L Giancardo, V Villemagne, C Rowe, P Yates, O Salvado and P Bourgeat. Computer-aided detection of cerebral microbleeds in susceptibility-weighted imaging. Computerized Medical Imaging and Graphics, 46, 269–276, 2015.
- Huang, C Michetti, M Busnelli, F Managò, S Sannino, D Scheggia, L Giancardo, D Sona, V Murino, B Chini, M L Scattoni and F Papaleo* “Chronic and Acute Intranasal Oxytocin Produce Divergent Social Effects in Mice”, Neuropsychopharmacology, 2014.
- L Giancardo*, D Sona, H Huang, S Sannino, F Managò, D Scheggia, F Papaleo and V Murino, “Automatic Visual Tracking and Social Behaviour Analysis with Multiple Mice”, PLOS ONE, 2013.
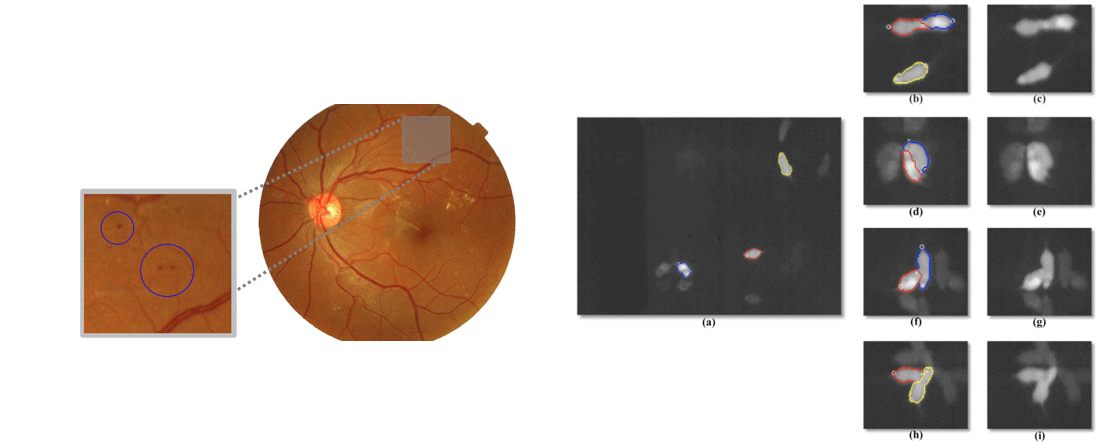
- L Giancardo*, F Meriaudeau, T P Karnowski, Y Li, S Gaarg, K W Tobin and E Chaum, “Exudate-based Diabetic Macular Edema Detection in Fundus Images Using Publicly Available Datasets”, Medical Image Analysis 16(1), 216—226, 2012.
- L Giancardo*, F Meriaudeau, T P Karnowski, K W Tobin, P Favaro, E Grisan, A Ruggeri and E Chaum, “Texture-less Macula Swelling Detection with Multiple Retinal Fundus Images”, IEEE Transactions on Biomedical Engineering, 2011.
 |
|
| Image/signal based computational biomarker development. |
Using modern machine learning approaches, we can have the opportunity of discovering data pattern from unstructured image or time signal data for developing new types of computational biomarkers for hypotheses generation or predicting outcomes. |
- Faculty/PI Name: Luca Giancardo
- Publications:
- L Giancardo*, K Roberts and Z Zhao, “Representation Learning for Retinal Vasculature Embeddings”. Fetal, Infant and Ophthalmic Medical Image Analysis. FIFI 2017, OMIA 2017. Lecture Notes in Computer Science, vol 10554. Springer, Cham, 2017.
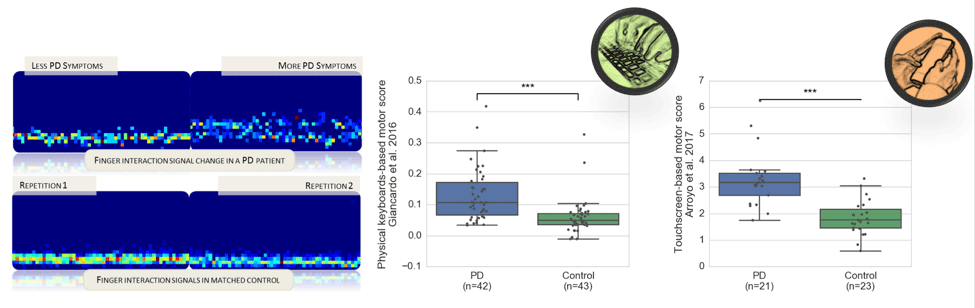
- T Arroyo-Gallego, M Ledesma-Carbayo, A Sanchez-Ferro, I Butterworth, C Mendoza, M Matarazzo, P Montero, R Lopez-Blanco, V Puertas-Martin, R Trincado and L Giancardo*. Detection of Motor Impairment in Parkinson’s Disease via Mobile Touchscreen Typing. IEEE Transaction on Biomedical Engineering, 64, 1994–2002, 2017.
- L Giancardo*, A Sanchez-Ferro, T. Arroyo-Gallego, I. Butterworth, C.S. Mendoza, P. Montero, M. Matarazzo, A. Obeso, M. L. Gray and San José Estepar, “Computer keyboard interaction as an indicator of early Parkinson's disease”, Scientific Reports, 6(34468), 2016.
- L Giancardo*, A Sanchez-Ferro, I Butterworth, C Sanchez-Mendoza and J M Hooker, “Psychomotor Impairment Detection via Finger Interactions with a Computer Keyboard”, Scientific Reports, 5(9678), 2015.
 |
|
| Genome3D Project |
Genome3D is a model-view framework for displaying genomic and epigenomic data within a three-dimensional physical model of the human genome. |
- Faculty/PI Name: Jim Zheng
- Publications:
- Asbury, T., Mitman, M., Tang, J., & Zheng, W. (2010). Genome3D: a viewer-model framework for integrating and visualizing multi-scale epigenomic information within a three-dimensional genome. BMC Bioinformatics, 11(1), 444. http://dx.doi.org/10.1186/1471-2105-11-444
- Website Link: http://www.genome3d.org/
|
|
| Ontology Fingerprints |
Ontology Fingerprint for a gene or a disease is a set of Gene Ontology terms overrepresented in the PubMed abstracts linked to a gene or disease along with those terms corresponding enrichment p-values. |
- Faculty/PI Name: Jim Zheng
- Publications:
- Tsoi, L., Boehnke, M., Klein, R., & Zheng, W. (2009). Evaluation of genome-wide association study results through development of ontology fingerprints. Bioinformatics, 25(10), 1314-1320. http://dx.doi.org/10.1093/bioinformatics/btp158
- Qin, T., Matmati, N., Tsoi, L., Mohanty, B., Gao, N., & Tang, J. et al. (2014). Finding pathway-modulating genes from a novel Ontology Fingerprint-derived gene network. Nucleic Acids Research, 42(18), e138-e138. http://dx.doi.org/10.1093/nar/gku678
- Website Link: http://www.ontologyfingerprint.org/
|
|
| RaPID |
RaPID is an ultra-fast tool for the identification of identity-by-descent segments among genotyped individuals. |
|
|
| HapSeq2 |
HapSeq2 is a program for genotyping calling and haplotype phasing from next generation sequencing data using haplotype information from jumping reads. |
|
|