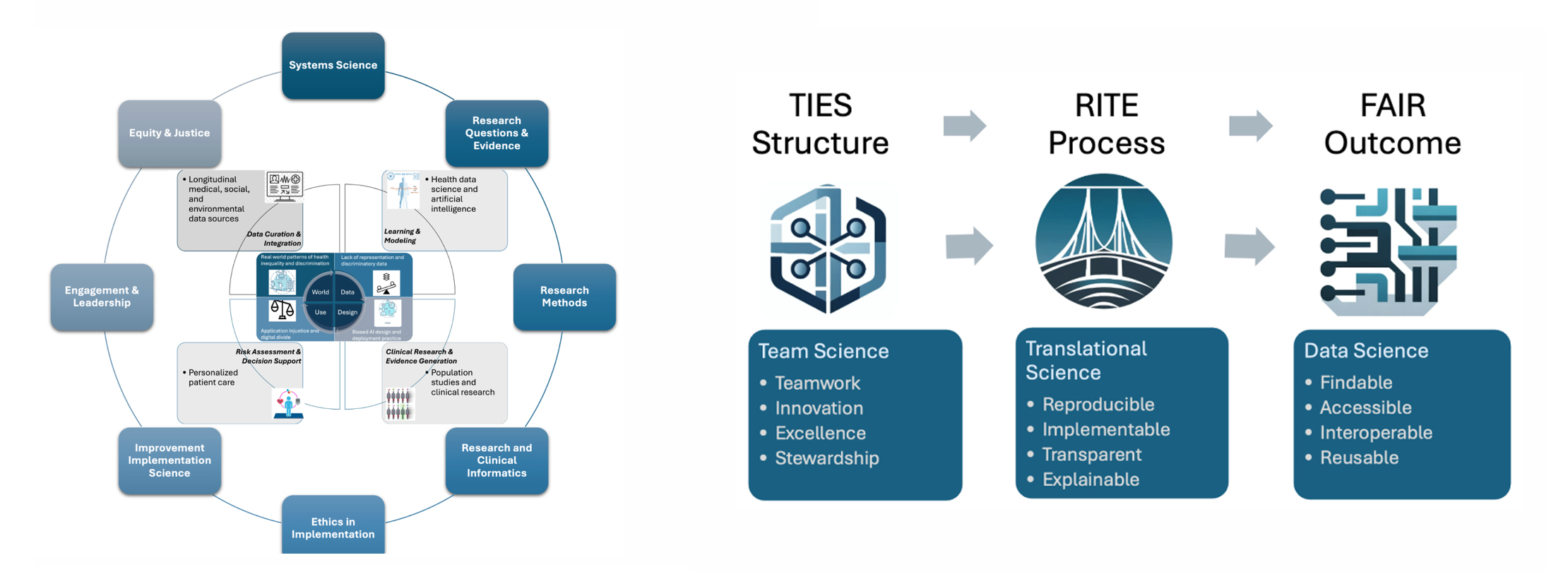
Learning health system (LHS) aims to leverage technology, data analytics, and evidence-based practices to create a feedback loop that continuously informs healthcare delivery, policy, and practice. It requires a multidisciplinary approach to interpret patterns observed in real-world data with the associated context. "A good structure increases the likelihood of good process, and good process increases the likelihood of good outcomes.” [Donabedian] We propose three guiding principles, as illustrated in the figure, to govern our research activities where we adopt team science approaches to achieve the translational goal of technology innovations developed based on real-world data.
Empowering these studies below:
The project is funded by Cancer Prevention & Research Institute of Texas (CPRIT) Established Investigator Award [RR230020].
Partnering with Rochester Epidemiology Project (REP) team at Mayo Clinic and Center for Spatial-temporal Modeling of Applications in Population Sciences (CSMAPS) at School of Public Health at UTHealth Houston, the Houston Epidemiology Linkage Project (HELP) aims to establish a record linkage system to empower studies of health and longevity across diverse populations leveraging longitudinal patient records (LPRs). Our goal is to adapt current geocoding, linkage, and follow-up methods used by the Rochester Epidemiology Project (REP) to facilitate studies of health and longevity across these diverse populations.
Intelligent Machine for Patient Accrual and Classification Tasks (IMPACT)
The project is funded by Cancer Prevention & Research Institute of Texas (CPRIT) Established Investigator Award [RR230020] and an NIH R01 grant “Semi-structured Information Retrieval in Clinical Text for Cohort Identification” [R01LM11934]
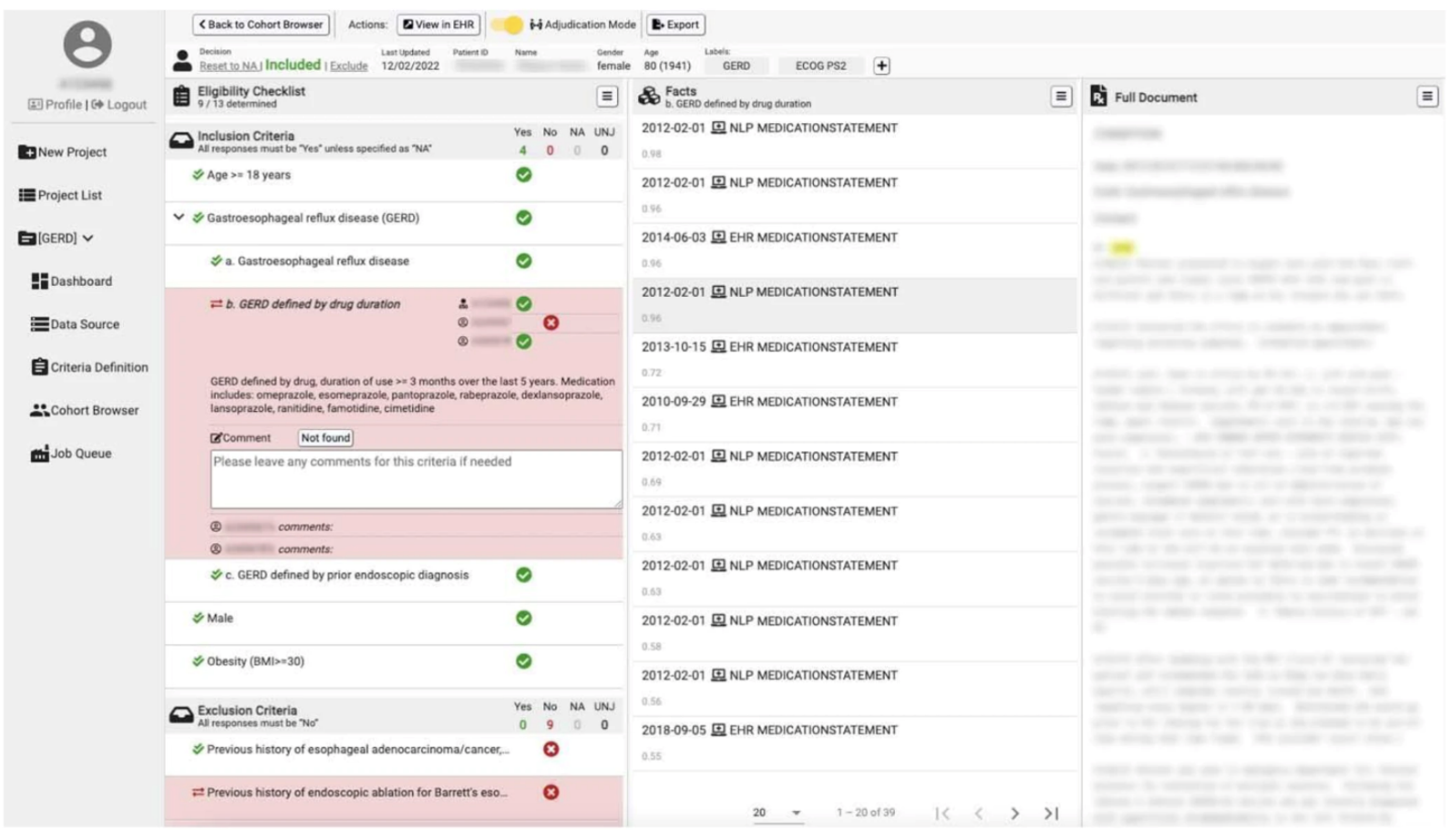
Clinical phenotyping is a critical task to the development of digital health applications in relation to the clinical domain. Clinical phenotype definitions often require information from multiple modalities of data beyond just structured EHR data (e.g., clinical notes, radiology images/interpretative reports, pathology samples/reports, etc.) and there is no guarantee all necessary data will be present/readily computationally accessible. We developed the IMPACT framework: a re-usable toolkit addressing challenges faced in silico clinical phenotyping.
Living Evidence Synthesis System (LIVES)
The project is funded by Cancer Prevention & Research Institute of Texas (CPRIT) Established Investigator Award [RR230020]
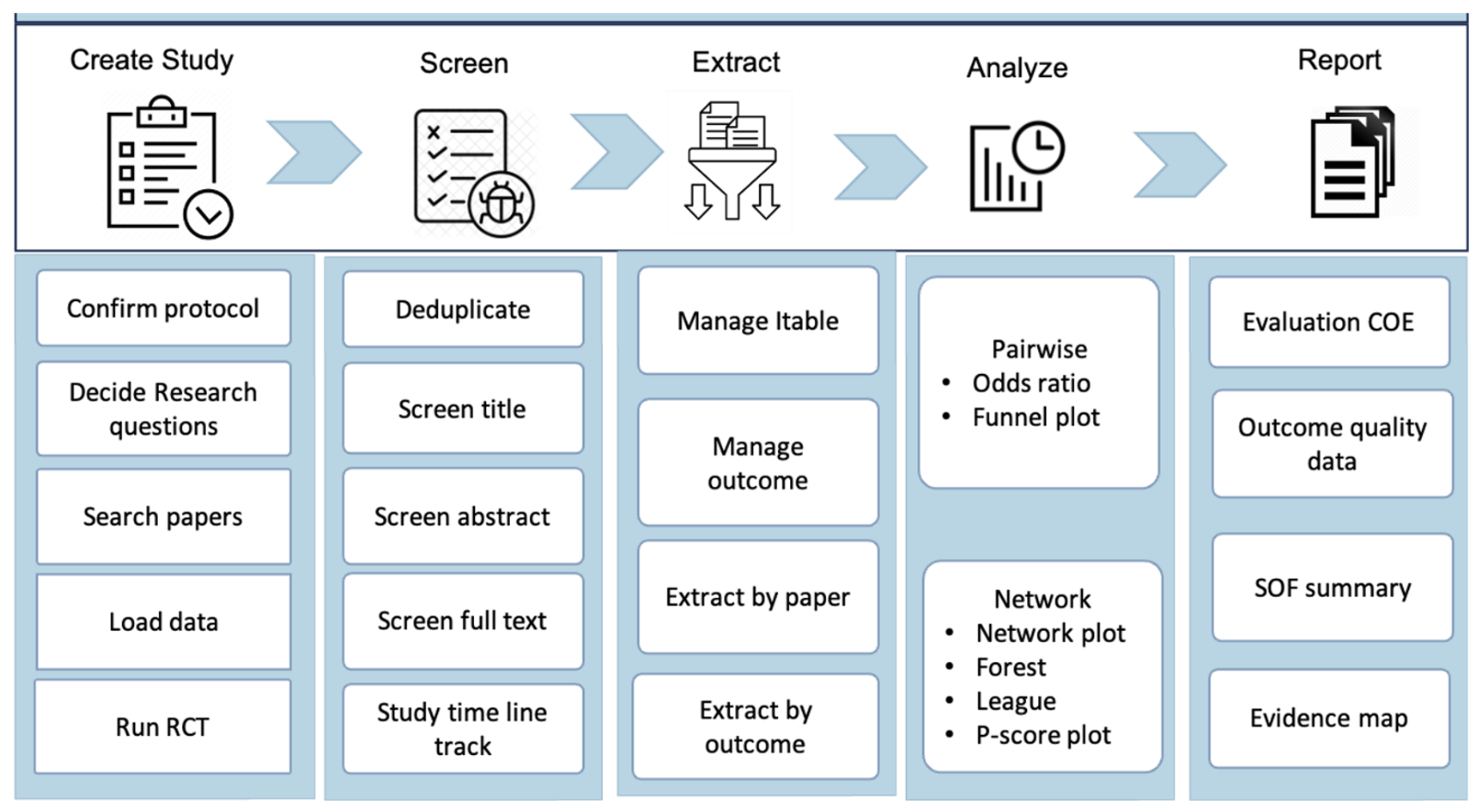
The task of evidence synthesis is to systematically gather, critically appraise, and summarize findings from multiple studies to provide a comprehensive overview of a particular topic or question. In clinical practice, popular evidence synthesis approaches include systematic reviews (SR) and meta-analyses (MA), aiming to synthesize evidence and provide decision makers with estimates of effect that are more precise than those provided by individual studies. Performing an SRMA study is time-intensive and resource-intensive process. As the conclusions become more outdated, can potentially lead to incorrect recommendations for clinical practice.
LIVES is a semi-automated AI-powered systematic review and meta-analysis platform aimed to ease the workload for researchers, while an adaptive PRISMA workflow and web-based platform empower experts to perform thorough systematic reviews. The platform is capable of handling intricate statistical techniques, including network or pairwise analysis, and producing standard outputs and decision aid for comparable evidence like certainty of evidence, summary of findings, and an evidence map. This system enables researchers to concentrate on achieving their research goals more efficiently.
Learning Precision Medicine for Rare Diseases Empowered by Knowledge-driven Data Mining [R01HG012748]
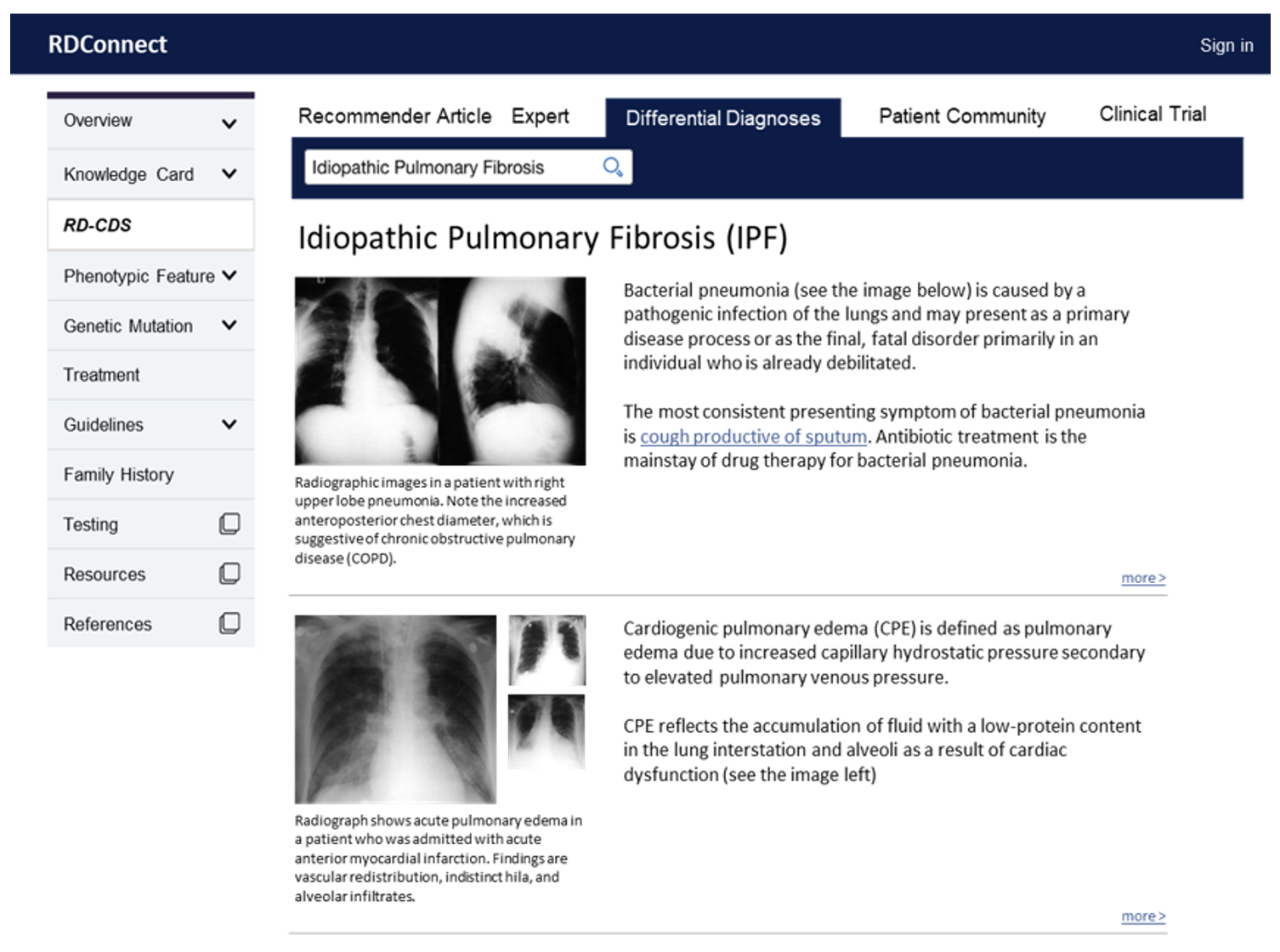
Despite their individual rarity, rare diseases collectively affect one in eleven Americans. Rare disease patients often face significant diagnostic delays, waiting an average of 6 years from the onset of symptoms to an accurate diagnosis. Recent advances in precision medicine have accelerated research in rare diseases, overwhelming clinicians’ capacities to manage and leverage the latest knowledge efficiently in clinical practice. Teaming up Mayo Clinic Program for Rare and Undiagnosed Diseases (PRaUD) with the partnership of Vanderbilt University Medical Center (VUMC), we aim to address the translation gap by building a novel end-to-end informatics framework to accelerate the diagnosis of rare diseases. The framework includes three components:
RDConnect - a web portal to search information, display differential diagnostic recommendations, and collect clinical evidence automatically for further validation.
Perioperative AI Research
The project is funded by NIH Grant “Secondary use of EHRs for Postsurgical Complication Surveillance” [R01EB19403].
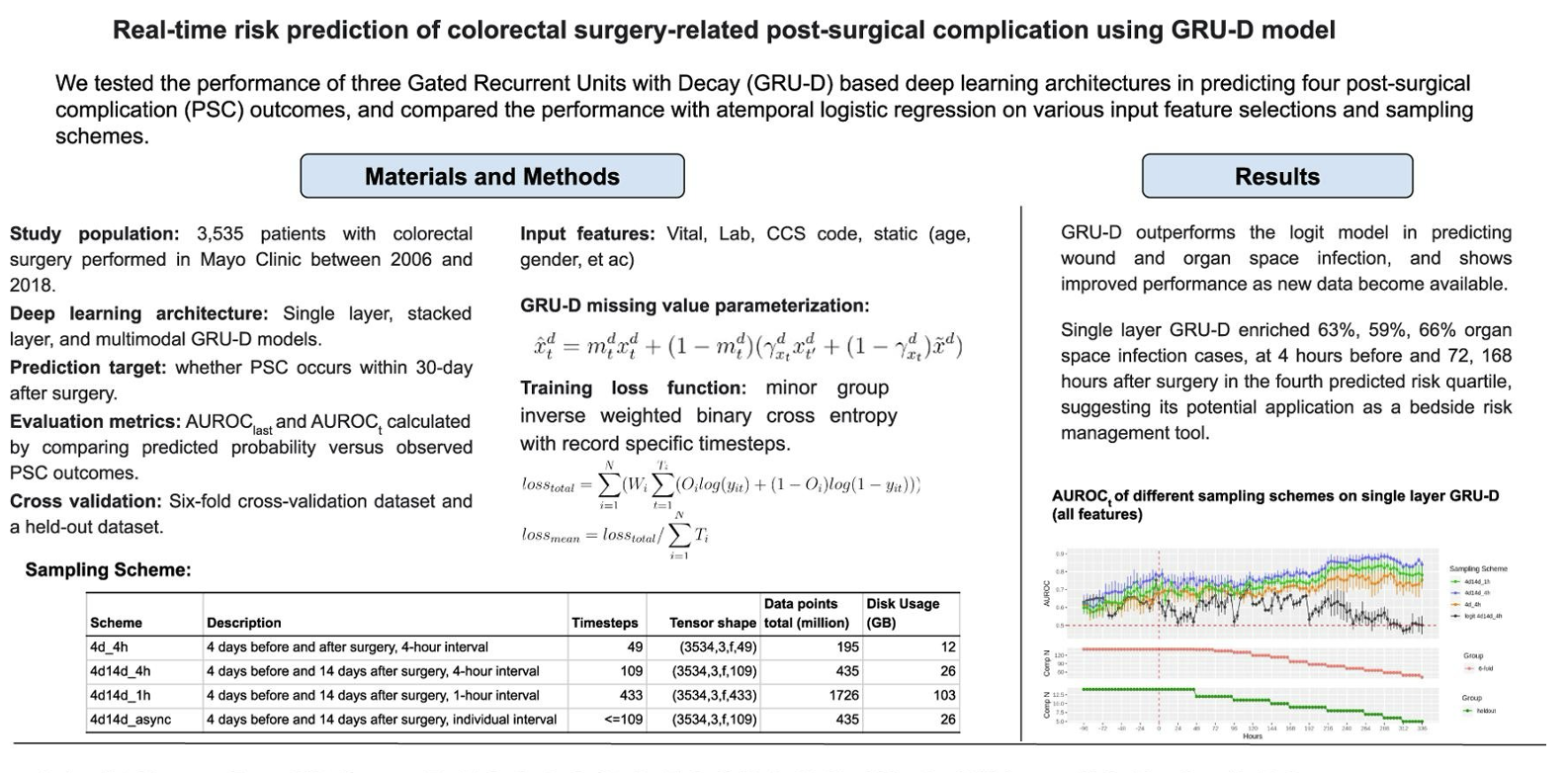
Partnering with Mayo Clinic and the Department of Anesthesiology, Critical Care, and Pain Medicine at the McGovern Medical School, we aim to develop and apply AI methods and tools to advance health care delivery, biomedical discovery, and public and population health in the perioperative setting.
An informatics framework for SUDEP Risk Marker Identification and Risk Assessment
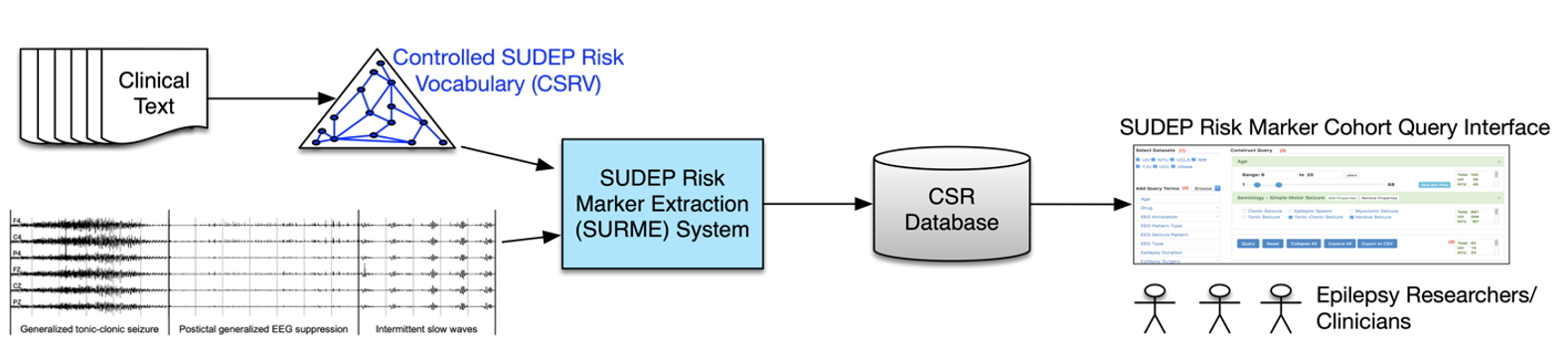
The project is funded by the National Institute of Neurological Disorders and Stroke (NINDS) [R01NS116287; PI: Licong Cui].
Sudden Unexpected Death in Epilepsy (SUDEP) remains a significant challenge in epilepsy care, claiming approximately 7,000 lives annually in the United States and Europe. The NINDS-funded Center for SUDEP Research (CSR), is dedicated to unraveling the biological mechanisms underlying SUDEP and developing interventions to prevent it. However, systematic assessment of SUDEP risk factors is hindered by fragmented and multimodal data sources (such as clinical text and electrophysiological signals), lack of standardized terminology for describing risk factors, and limited computational tools for extracting risk markers from multimodal data sources. To address these challenges, we propose to develop a controlled vocabulary for SUDEP risk factors and a SUDEP Risk Marker Extraction (SURME) system. Leveraging the multidimensional CSR data repository encompassing over 2,500 patients from 7 medical centers, SURME aims to automate the extraction of known and potential SUDEP risk markers from multimodal data sources.
CAREER: Advancing the Role of Ontologies for Data Science in Biomedicine
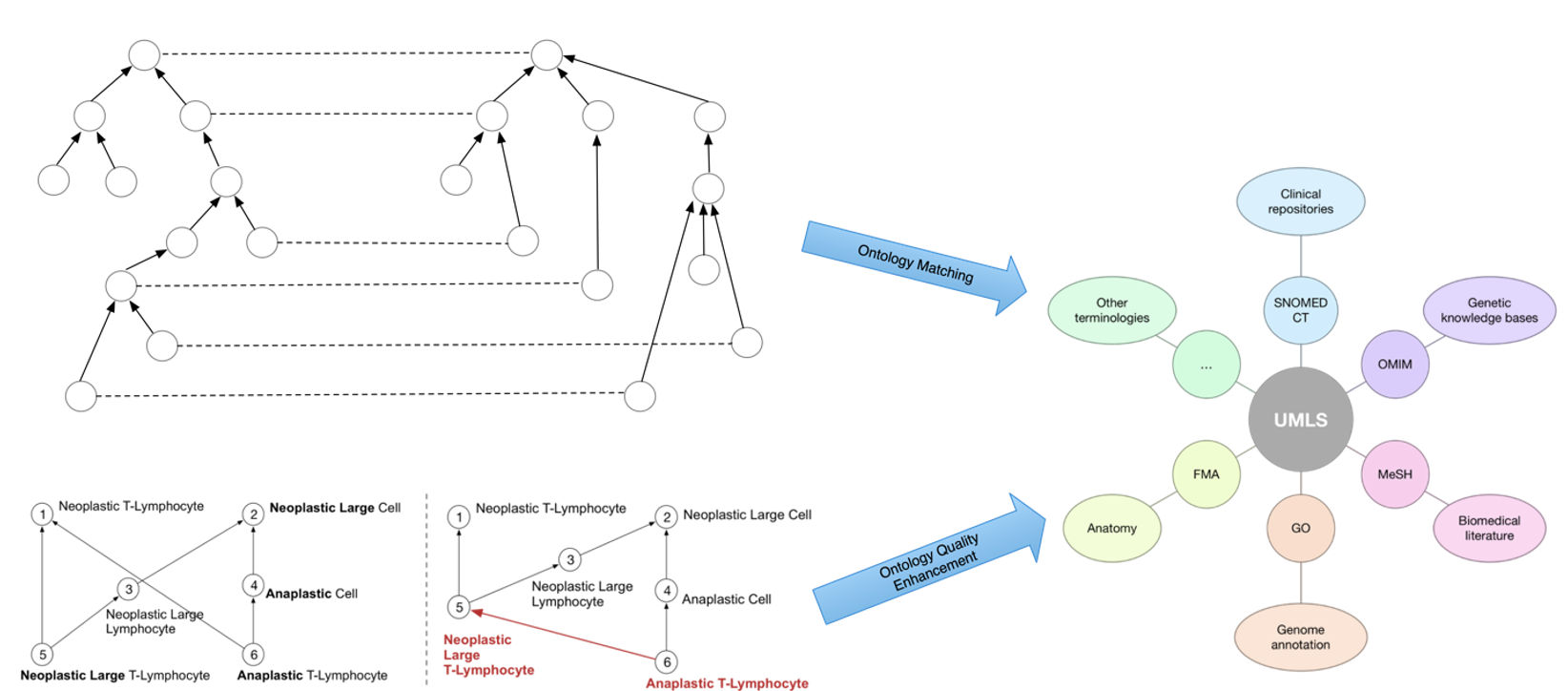
The project is funded by the National Science Foundation (NSF) [2047001; PI: Licong Cui].
Ontologies and terminologies have played a vital role in biomedical research for coding, managing, sharing, and exchange of vast amounts of heterogeneous biomedical data that are being continuously generated, such as in Electronic Health Records (EHRs). EHRs have been widely used in translational research to learn predictive models for discovery and disease management across varying patient cohorts. The very first step in such EHR-based applications often concerns patient cohort identification. However, there are two critical barriers in performing effective cohort identification from large-scale EHRs. The first one is data (or semantic) heterogeneity, caused by a mixed utilization of coding systems. The second one is the quality of the semantic backbone or ontology hierarchy, which is essential for translating patient eligibility criteria to executable database queries. To address such challenges, this project aims to develop new deep learning-based methods for ontology matching and for ontology quality enhancement that directly impact data science practice in biomedicine, such as patient cohort identification.