
Assistant Professor
Department of Bioinformatics and Systems Medicine
Contact
[email protected] | 713-500-3698
Assaf Gottlieb, Ph.D., joined McWilliams School of Biomedical Informatics at UTHealth Houston, formerly UTHealth Houston School of Biomedical Informatics (SBMI) in January 2017 as an assistant professor and one of the core faculty members of the Center for Precision Health.
Dr. Gottlieb arrived to McWilliams School of Biomedical Informatics from IBM Research where he served as a research associate at the Machine Learning for Healthcare and Life Science group in Haifa labs. Prior to his position with IBM, Dr. Gottlieb was a postdoctoral fellow at both Stanford University and Tel-Aviv University. He has degrees in Physics and Computer Science and has also worked in software and algorithmic development in various software companies.
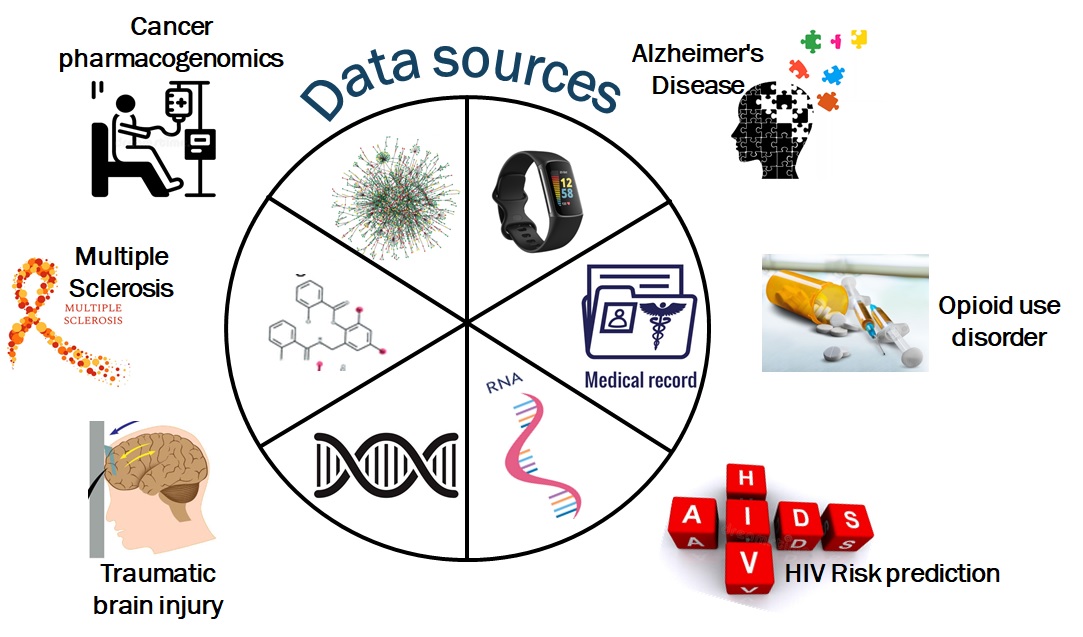
Our lab is interested in precision medicine with special emphasis on pharmacogenomics. In order to address the challenges of precision medicine, we employ an integrative computational approach, combining multiple types of data and applying methodologies from systems biology, systems medicine, machine learning and causal inference domains. Our primary interest is to improve health outcomes by integrating diverse types of data, including genomic/biological data, data from fitness trackers and clinical data from electronic health records data.
Some of our on-going projects include predicting drug response in cancer therapy, early predictions of Alzheimer’s Disease and determining HIV risk.
You can find the full set of publications in NCBI Bibliography or Google Scholar in the top right corner. Below is a selected set from recent years.